Phospholipids and cholesterol are both important components of the cell membrane. They play an integral role in cell transmembrane transport, signal transmission, and biological metabolism. But what are their interactions? In this literature review, we will share an article on the analysis of the interactions between phospholipids and cholesterol (CHOL) using two means: scattering and molecular dynamics simulation. It is hoped that it will not only improve our understanding of cell membrane structure, but also bring inspirations to the development of liposomes and lipid nanoparticles.
 Abstract:
Abstract:
Cholesterol and phospholipids are ubiquitous in mammalian cell membranes and their interactions are critical in lipid-mediated cholesterol trafficking. The interactions between cholesterol and phospholipids were determined by combining small-angle neutron scattering (SANS), small-angle X-ray scattering (SAXS), and molecular dynamics (MD) simulation of the whole atom.
Modeling: The scattering density distribution model of bilayers composed of cholesterol and ether lipids was constructed, and the structural parameters of various bilayer membranes were obtained.
Simulation: Molecular dynamics simulations with surface area constraints were performed and the experimental data were reproduced. This iterative analytical approach yielded good agreement between the experimental and simulated structures, and molecular interactions between cholesterol and phospholipids were further verified by MD simulations.
Conclusion: · In the presence of ether lipid, the hydroxyl group (-OH) of cholesterol mainly forms hydrogen bonds with the phosphate group on the phospholipid head. In the presence of phospholipid, the hydroxyl group of cholesterol forms hydrogen bonds with the carbonyl group on the phospholipid fat chain. In summary, cholesterol moves closer to the bilayer surface when ether lipids are present and prompts dehydration of phosphate groups in the head. In addition, the three-dimensional spatial density distribution around cholesterol suggests that it undergoes anisotropic chain packing, resulting in tilt of cholesterol.
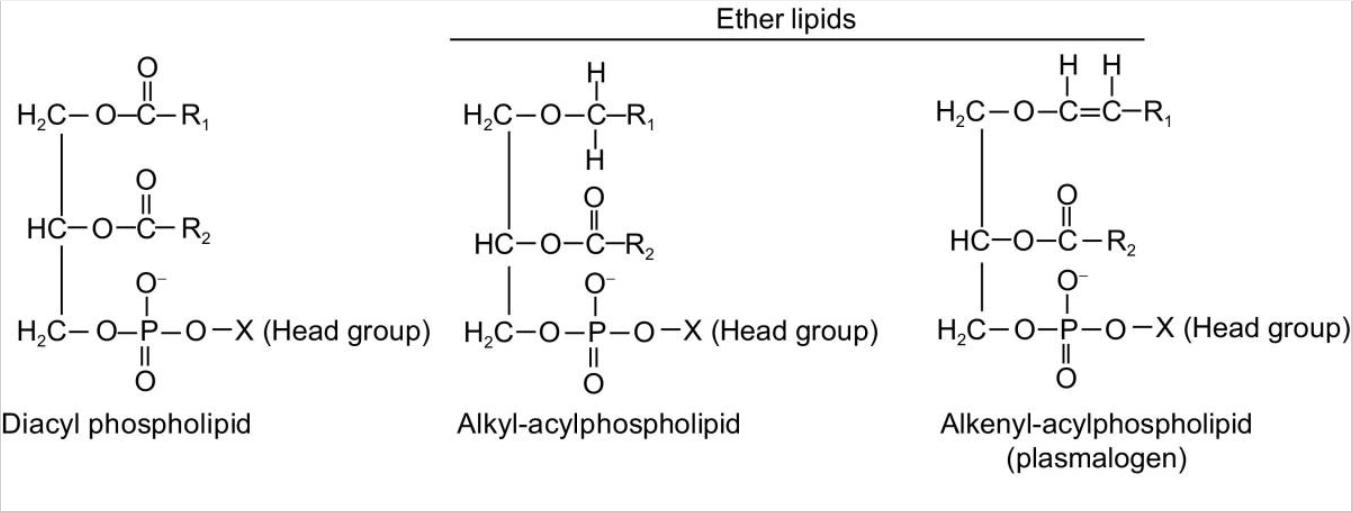
Fig. 1. Molecular structurs of phospholipids and ether lipids
Material:
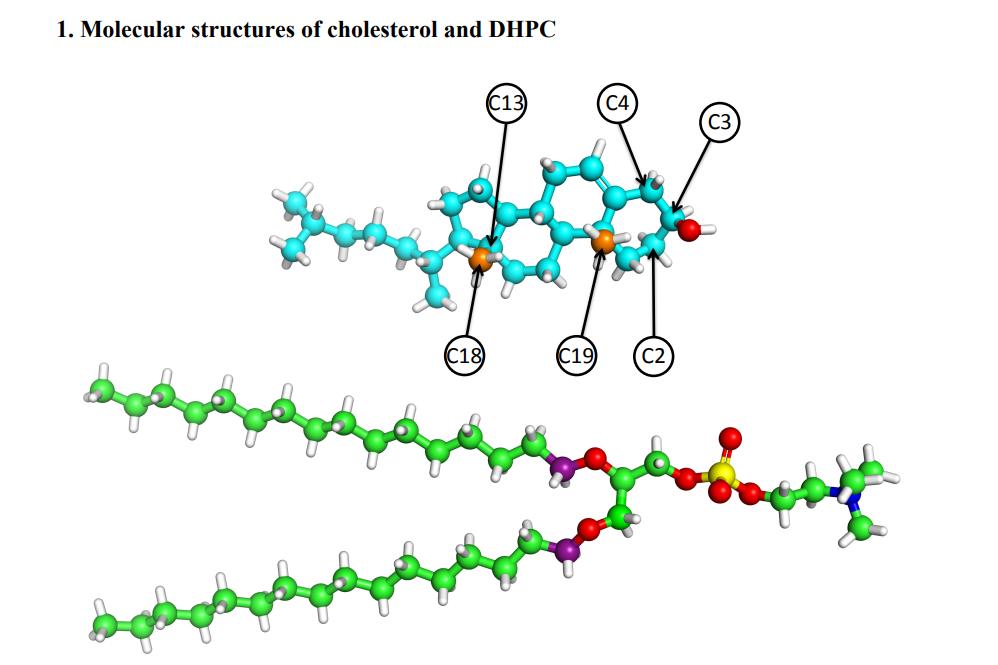
Fig. 2. Molecular structures of cheolesterol (CHOL) and dihexadecylphosphatidylcholine (DHPC)
The constructed model is a lipid bilayer composed of cholesterol (CHOL) and dihexadecylphosphatidylcholine (DHPC). Atomic images of the scattering density distribution (SDP) model were first obtained by MD simulation without applying surface tension, then different structural parameters (lipid area, bilayer hydrophobic layer thickness, and so on) were determined, and the model was then optimized by analyzing the experimental data. MD simulation was performed upon the surface tension at the later stage, and the molecular interactions between cholesterol and different lipids were further analyzed by comparing the simulation results with the test results.
Small-angle neutron scattering (SANS):
Neutron testing was performed using the Scattered Neutron Source (SNS) BL-6 EQ-SANS at Oak Ridge National Laboratory. The total scattering vector Q (0.03 < Q < 0.8 Å -1) was calculated using a detector at a range of wavelengths (2.5-6.0 Å), with a distance from the detector to sample of 2.0 m. Using software provided by SNS for correction and noise reduction, one-dimensional (1D) intensities were obtained by data analysis.
Small-angle X-ray scattering (SAXS):
X-ray data were collected from the Cornell High Energy Synchrotron Source G-1. The charge coupler FLICAM (71 µm linear size, pixel array of 1,024 Í 1,024) was used to detect scattering from a collimated incident beam (0.24 Í 0.24 mm2) illuminated by X-rays at a wavelength of 1.17 Å on ULV samples. The distance from sample to detector was 505.8 mm. The sample was transferred into a 1.5-mm quartz capillary tube and placed in a temperature-controlled rack. The relationship between scattering intensity I and scattering vector Q was obtained by background subtracting radial averaging of two-dimensional (2D) data. It is then converted to an X-ray shape factor using the same relationship as neutrons.
Molecular Dynamics Simulation (MD):
CHARMM-GUI Membrane Builder was used to generate coordinates for a pure DHPC bilayer (128 DHPC) and a DHPC bilayer with 20 mol% cholesterol (128DHPC + 32CHOL).
Because the ether lipid DHPC was not included in the lipid selection of CHARMM-GUI, it was necessary to firstly build the DPPC bilayer model using CHARMM-GUI, and then change the carbonyl group (C = O) in DPPC to methylene group (CH2) in DHPC. The system was solvated by adding 4,300-4,800 water molecules, while sufficient NaCl was introduced for charge balance. The NAMD package (version 2.816) and CHARMM 36 lipid force field were used for MD simulations. For specific force field optimization and MD parameter settings, refer to the original text as well as supporting documents.
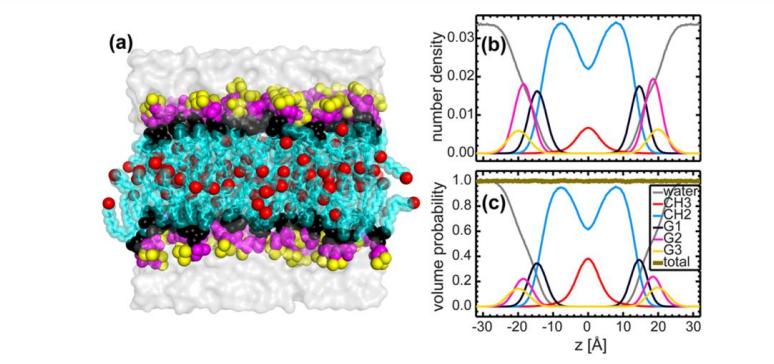
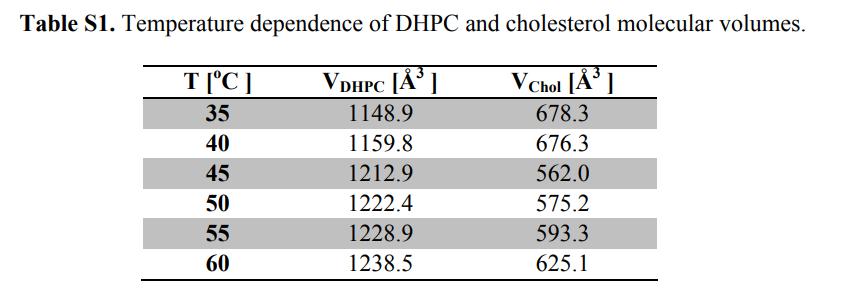
Fig. 3. MD simulation system and DHPC bilayer system construction
(a) Equilibrated bilayer systems in which the lipid bilayer is resolved into five components (color differentiation): red-methyl (CH3); black-glycerol backbone + ether bond; light-blue-methylene (CH2); purple-head group (phosphate radical + CH2CH2N); and yellow-choline terminal trimethyl.
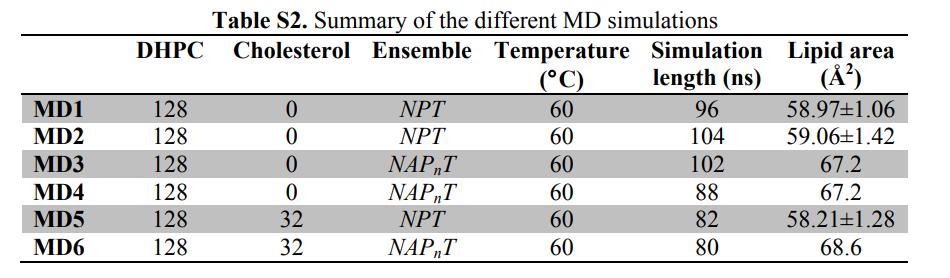
Fig. 4. Elucidation of SDP models by different comparative experimental factors
(The lipid composition is described by analytical functions (i.e., Gaussian function and error function), and the water content is determined by complementation with the total liposome integral. a: neutron scattering; b: x-ray scattering; c: total electron density; d: neutron scattering length density; e: volume distribution. Note: Color description is consistent with Fig. 3)
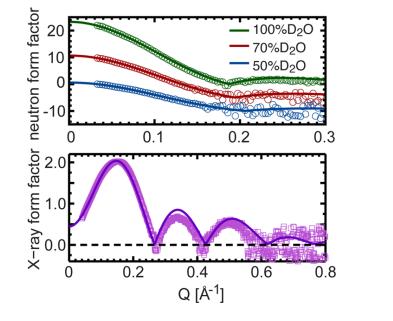
Fig. 4. Direct comparison between neutron and X-ray shape factors for different contrast experiments
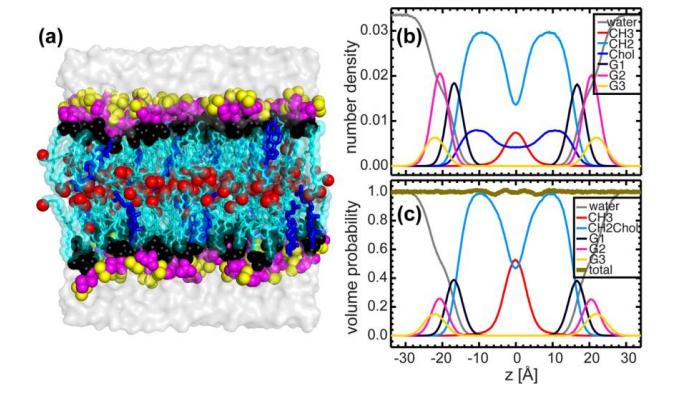
Fig. 5. MD simulation of a bilayer containing 20 mol% cholesterol and DHPC
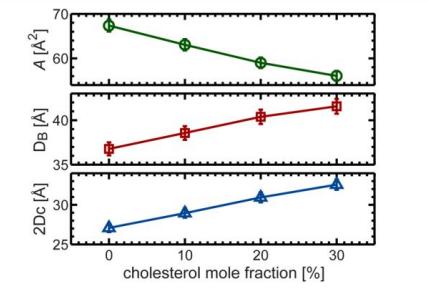
Fig. 6. Concentration of cholesterol versus changes in representative structural parameters for DHPC bilayers
(The ordering effect of cholesterol is demonstrated by a decrease in surface lipid area with increasing cholesterol concentration, an increase in lipid membrane bilayer thickness DB and hydrocarbon thickness 2DC, and an approximately linear change in the aggregation effect of cholesterol.)
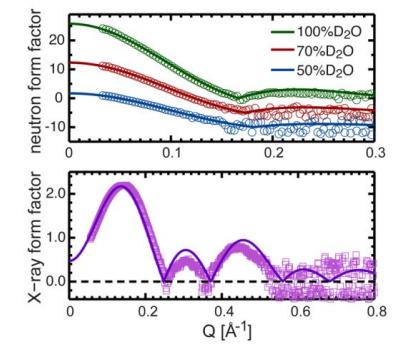
Fig. 7. Direct comparison between different experimental form factors for DHPC bilayer membranes consisting of 20 mol% cholesterol and form factors calculated by NAPnT Kinetic Simulation at the fixed unit cell area
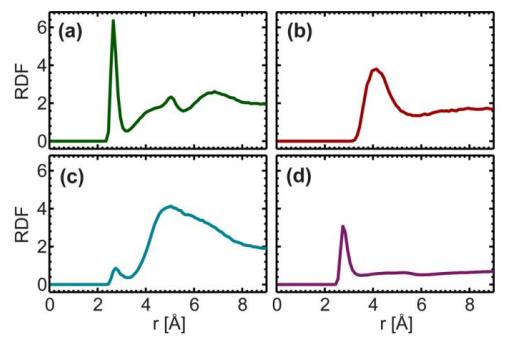
Fig. 8. Radial distribution function of hydroxyl group (-OH) of cholesterol with phosphate radical (a), N atom of choline (b), main chain ether group (-O-, c), and oxygen atom in free water (A sharp peak was observed for phosphate radical and water at 2.7 Å, and a broad peak was observed for N atom in choline at 4 Å.)
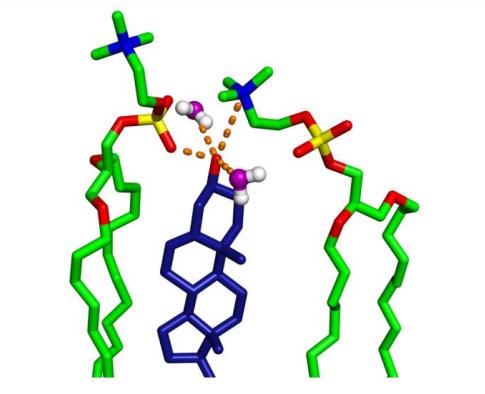
Fig. 9. Hydrogen bonding between CHOL-OH and nearby lipids based on the RDF distribution in Fig. 8. (Orange indicates potential hydrogen bonds and electrostatic interactions.)
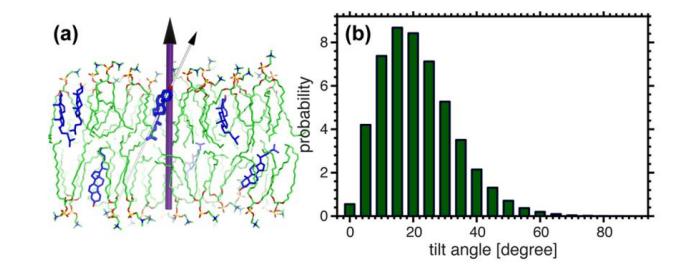
Fig. 10. Obliquity of the planar tetraloop of cholesterol relative to the normal of the bilayer. Cholesterol is not vertically distributed in the membrane, but with a certain angle. The vast majority of cholesterol has a 20° tilt distribution.
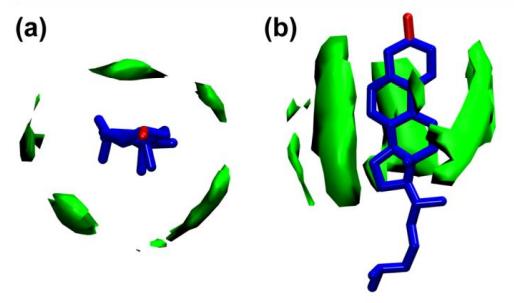
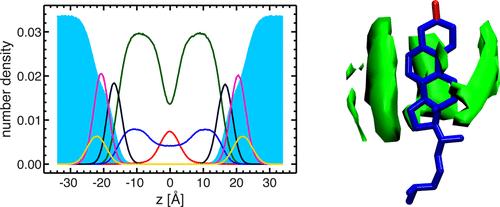
Fig. 11. 3D density distribution of aliphatic hydrocarbon chains near cholesterol. Anisotropic chain packing is associated with cholesterol tetraloops, and high-density chain packing also occurs near the smooth face of the ring.
Conclusion:
This study combined experiments with simulations. SANS and SAXS were used to investigate the molecular position and orientation of cholesterol in ether lipid bilayers. The causes for different scattering properties of bilayers were further analyzed by MD simulations, which formed the basis of the SDP model for analyzing the experimental data. In contrast, the model parameters were further optimized using the structural parameters obtained experimentally to improve the authenticity and reliability of MD simulations and determine molecular interactions between cholesterol and ether lipids. Molecular interactions between ordinary acylphospholipids and cholesterol are quite another.
Specifically:
In bilayers composed of ether lipids, the hydroxyl group (-OH) on cholesterol forms hydrogen bonds with oxygen in phosphate groups, while in ordinary phospholipids, hydroxyl groups tend to form hydrogen bonds with carbonyl groups (C = O) on phospholipid fats due to the presence of ester groups. This difference in the interactions may explain the important role that acetal phospholipids (ether lipids) play in mediating cholesterol transport. More importantly, in the present study, the interactions between different molecules in the membrane environment were further modulated by changing the chemical composition of the lipids.
Cholesterol, including cholesterol HP, plays a great role in organisms and is also an important pharmaceutical excipient. AVT supplies plant-derived cholesterol. Contact us for more information.
References:
[1]. Dean JM, Lodhi IJ. Structural and functional roles of ether lipids. Protein Cell. 2018;9(2):196-206. doi:10.1007/s13238-017-0423-5
[2]. Pan J, Cheng X, Heberle FA, Mostofian B, Kučerka N, Drazba P, Katsaras J. Interactions between ether phospholipids and cholesterol as determined by scattering and molecular dynamics simulations. J Phys Chem B. 2012 Dec 27;116(51):14829-38. doi: 10.1021/jp310345j. Epub 2012 Dec 13. PMID: 23199292; PMCID: PMC3539752.